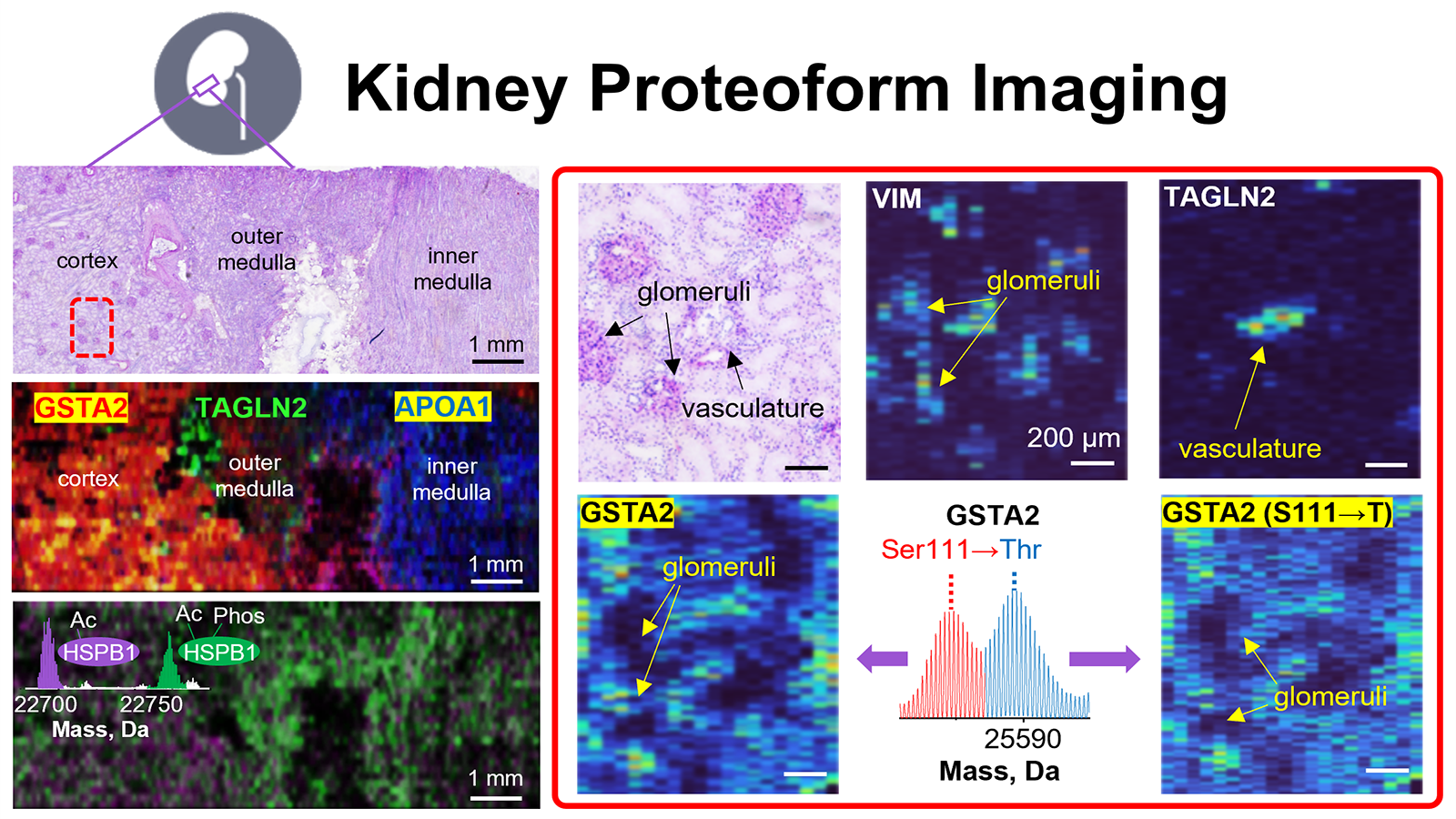
The human body is made up of trillions of cells, each having a specialized role, and each affected by its environment and neighboring cells. The NIH Common Fund’s Human BioMolecular Atlas Program (HuBMAP) is a consortium of over 300 researchers using cutting-edge techniques to study the spatial organization of cells within tissues to better understand normal function of organs in health and disease. One kind of these techniques is called “proteomics” which studies the proteins within each of the cells that make up the body. By knowing which proteins are within specific cell types, researchers can better determine the types of cells that make up an organ, and where those cells are within that organ, which will assist researchers in finding and treating disease. While studying proteins is a good way to identify cells within a tissue, proteins can have different types of molecules attached to them that change their function and activity. When these molecules are added to a protein, it becomes a “proteoform,” and one protein can have many proteoforms, depending on the types of molecules that have been added. Since the proteoforms are different versions of the same protein, they are a more specific marker of cell type and function, and thus it is vital for researchers to develop new technologies that can identify the multitudes of proteoforms within a tissue section. To this end, Neil Kelleher, Ph.D., and his group at Northwestern published “Highly multiplexed, label-free proteoform imaging of tissues by individual ion mass spectrometry” in the August 2022 issue of Science Advances, describing the use of Proteoform Image Mass Spectrometry (PiMS) to identify and image proteoforms within slices of human kidney. PiMS combines two different types of high-throughput techniques called proteomics to determine the mass of the proteoform, and to create an image of that proteoform within the tissue. With the information gathered from these methods, the researchers then searched a custom-made database of 1000 proteoforms that are known to be in the kidney to identify the proteoforms they found. Once they knew what proteoforms they were looking at, they were able to make one single composite image of the location of many proteoforms within the same section of kidney, like the one below. PiMS is able to determine the types of proteoforms within the kidney section at a spatial resolution of 80 micrometers, allowing researchers insights into the proteins that comprise human tissues at a level not seen before. PiMS can identify and reveal the location of proteoforms within an organ, making it a promising new application for tissue mapping efforts, biomarker discovery, and disease diagnostics, as well as being able to unveil what was once unseen.
Highly multiplexed, label-free proteoform imaging of tissues by individual ion mass spectrometry
Pei Su 1, John P McGee 1, Kenneth R Durbin 1, Michael A R Hollas 1, Manxi Yang 2, Elizabeth K Neumann 3, Jamie L Allen 3, Bryon S Drown 1, Fatma Ayaloglu Butun 4, Joseph B Greer 1, Bryan P Early 1, Ryan T Fellers 1, Jeffrey M Spraggins 3 5, Julia Laskin 2, Jeannie M Camarillo 1 4, Jared O Kafader 1 4, Neil L Kelleher 1 4 6
Sci Adv. 2022 Aug 12;8(32):eabp9929. doi: 10.1126/sciadv.abp9929. Epub 2022 Aug 10.
PMID: 35947651 PMCID: PMC9365283 DOI: 10.1126/sciadv.abp9929
This work is supported by NIH Common Fund grant UH3CA246635.


